library(rtemis)2 rtemis in 60 seconds
2.1 Load rtemis
2.2 Regression
For regression, the outcome must be continuous
x <- rnormmat(500, 50, seed = 2019)
w <- rnorm(50)
y <- x %*% w + rnorm(500)
dat <- data.frame(x, y)
res <- resample(dat)05-20-25 07:06:21 Input contains more than one columns; will stratify on last :resample
.:Resampling Parameters
n.resamples: 10
resampler: strat.sub
stratify.var: y
train.p: 0.75
strat.n.bins: 4
05-20-25 07:06:21 Created 10 stratified subsamples :resample
dat.train <- dat[res$Subsample_1, ]
dat.test <- dat[-res$Subsample_1, ]2.2.1 Check Data
check_data(x) x: A data.table with 500 rows and 50 columns
Data types
* 50 numeric features
* 0 integer features
* 0 factors
* 0 character features
* 0 date features
Issues
* 0 constant features
* 0 duplicate cases
* 0 missing values
Recommendations
* Everything looks good
2.2.2 Single Model
mod <- s_GLM(dat.train, dat.test)05-20-25 07:06:21 Hello, egenn :s_GLM
.:Regression Input Summary
Training features: 374 x 50
Training outcome: 374 x 1
Testing features: 126 x 50
Testing outcome: 126 x 1
05-20-25 07:06:21 Training GLM... :s_GLM
.:GLM Regression Training Summary
MSE = 1.02
RMSE = 1.01
MAE = 0.81
r = 0.99 (p = 1.3e-310)
R sq = 0.98
.:GLM Regression Testing Summary
MSE = 0.98
RMSE = 0.99
MAE = 0.76
r = 0.99 (p = 2.7e-105)
R sq = 0.98
05-20-25 07:06:21 Completed in 3.2e-04 minutes (Real: 0.02; User: 0.02; System: 1e-03) :s_GLM
2.2.3 Crossvalidated Model
mod <- train_cv(dat, mod = "glm")05-20-25 07:06:21 Hello, egenn :train_cv
.:Regression Input Summary
Training features: 500 x 50
Training outcome: 500 x 1
05-20-25 07:06:21 Training Ranger Random Forest on 10 stratified subsamples... :train_cv
05-20-25 07:06:21 Outer resampling plan set to sequential :resLearn
.:Cross-validated Ranger
Mean MSE of 10 stratified subsamples: 27.48
Mean MSE reduction: 44.11%
05-20-25 07:06:24 Completed in 0.04 minutes (Real: 2.26; User: 12.21; System: 0.23) :train_cv
Use the describe function to get a summary in (plain) English:
mod$describe()Regression was performed using Ranger Random Forest. Model generalizability was assessed using 10 stratified subsamples. The mean R-squared across all testing set resamples was 0.44.mod$plot()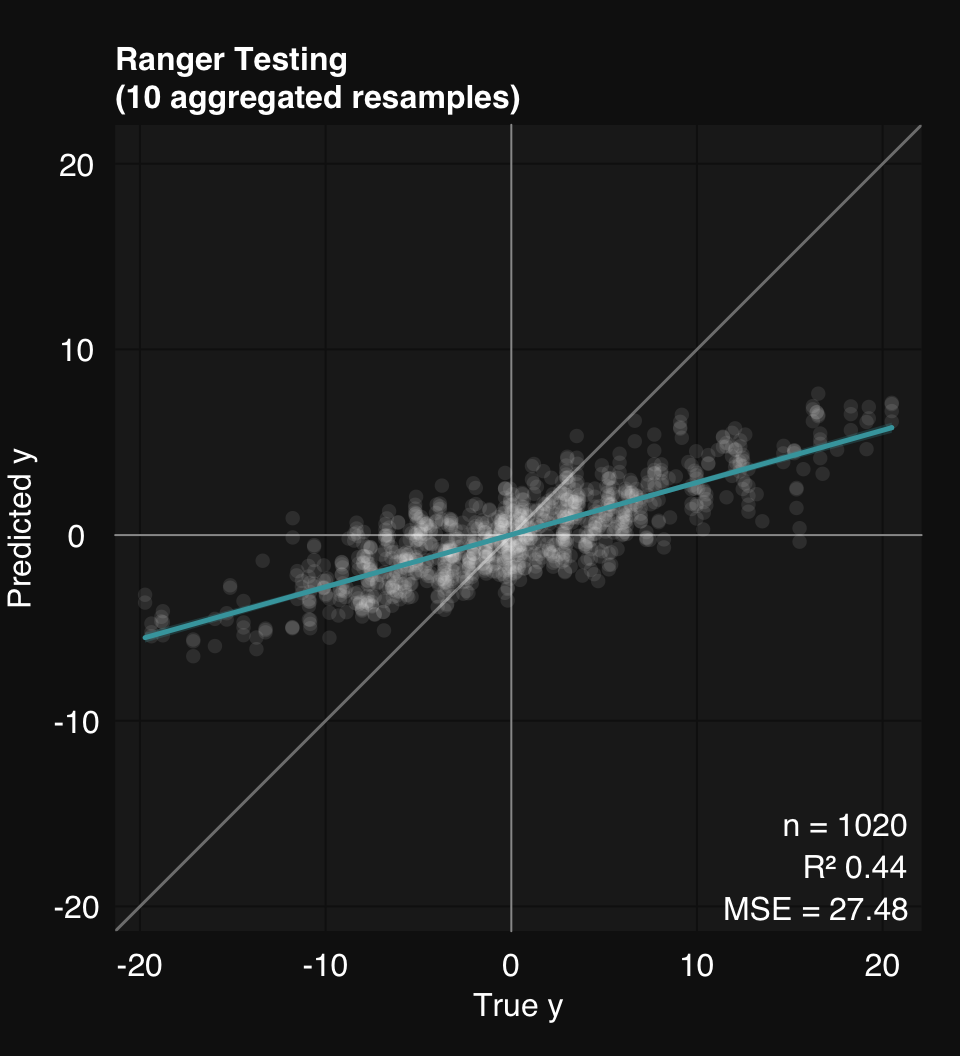
2.3 Classification
For classification the outcome must be a factor. In the case of binary classification, the first level should be the “positive” class.
2.3.1 Check Data
data(Sonar, package = 'mlbench')
check_data(Sonar) Sonar: A data.table with 208 rows and 61 columns
Data types
* 60 numeric features
* 0 integer features
* 1 factor, which is not ordered
* 0 character features
* 0 date features
Issues
* 0 constant features
* 0 duplicate cases
* 0 missing values
Recommendations
* Everything looks good
res <- resample(Sonar)05-20-25 07:06:24 Input contains more than one columns; will stratify on last :resample
.:Resampling Parameters
n.resamples: 10
resampler: strat.sub
stratify.var: y
train.p: 0.75
strat.n.bins: 4
05-20-25 07:06:24 Using max n bins possible = 2 :strat.sub
05-20-25 07:06:24 Created 10 stratified subsamples :resample
sonar.train <- Sonar[res$Subsample_1, ]
sonar.test <- Sonar[-res$Subsample_1, ]2.3.2 Single model
mod <- s_Ranger(sonar.train, sonar.test)05-20-25 07:06:24 Hello, egenn :s_Ranger
05-20-25 07:06:24 Imbalanced classes: using Inverse Frequency Weighting :prepare_data
.:Classification Input Summary
Training features: 155 x 60
Training outcome: 155 x 1
Testing features: 53 x 60
Testing outcome: 53 x 1
.:Parameters
n.trees: 1000
mtry: NULL
05-20-25 07:06:24 Training Random Forest (ranger) Classification with 1000 trees... :s_Ranger
.:Ranger Classification Training Summary
Estimated
Reference M R
M 83 0
R 0 72
Overall
Sensitivity 1.0000
Specificity 1.0000
Balanced Accuracy 1.0000
PPV 1.0000
NPV 1.0000
F1 1.0000
Accuracy 1.0000
AUC 1.0000
Brier Score 0.0176
Positive Class: M
.:Ranger Classification Testing Summary
Estimated
Reference M R
M 25 3
R 11 14
Overall
Sensitivity 0.8929
Specificity 0.5600
Balanced Accuracy 0.7264
PPV 0.6944
NPV 0.8235
F1 0.7812
Accuracy 0.7358
AUC 0.8643
Brier Score 0.1652
Positive Class: M
05-20-25 07:06:24 Completed in 1.2e-03 minutes (Real: 0.07; User: 0.16; System: 0.03) :s_Ranger
2.3.3 Crossvalidated Model
mod <- train_cv(Sonar)05-20-25 07:06:24 Hello, egenn :train_cv
.:Classification Input Summary
Training features: 208 x 60
Training outcome: 208 x 1
05-20-25 07:06:24 Training Ranger Random Forest on 10 stratified subsamples... :train_cv
05-20-25 07:06:24 Outer resampling plan set to sequential :resLearn
.:Cross-validated Ranger
Mean Balanced Accuracy of 10 stratified subsamples: 0.83
05-20-25 07:06:24 Completed in 0.01 minutes (Real: 0.70; User: 1.63; System: 0.15) :train_cv
mod$describe()Classification was performed using Ranger Random Forest. Model generalizability was assessed using 10 stratified subsamples. The mean Balanced Accuracy across all testing set resamples was 0.83.mod$plot()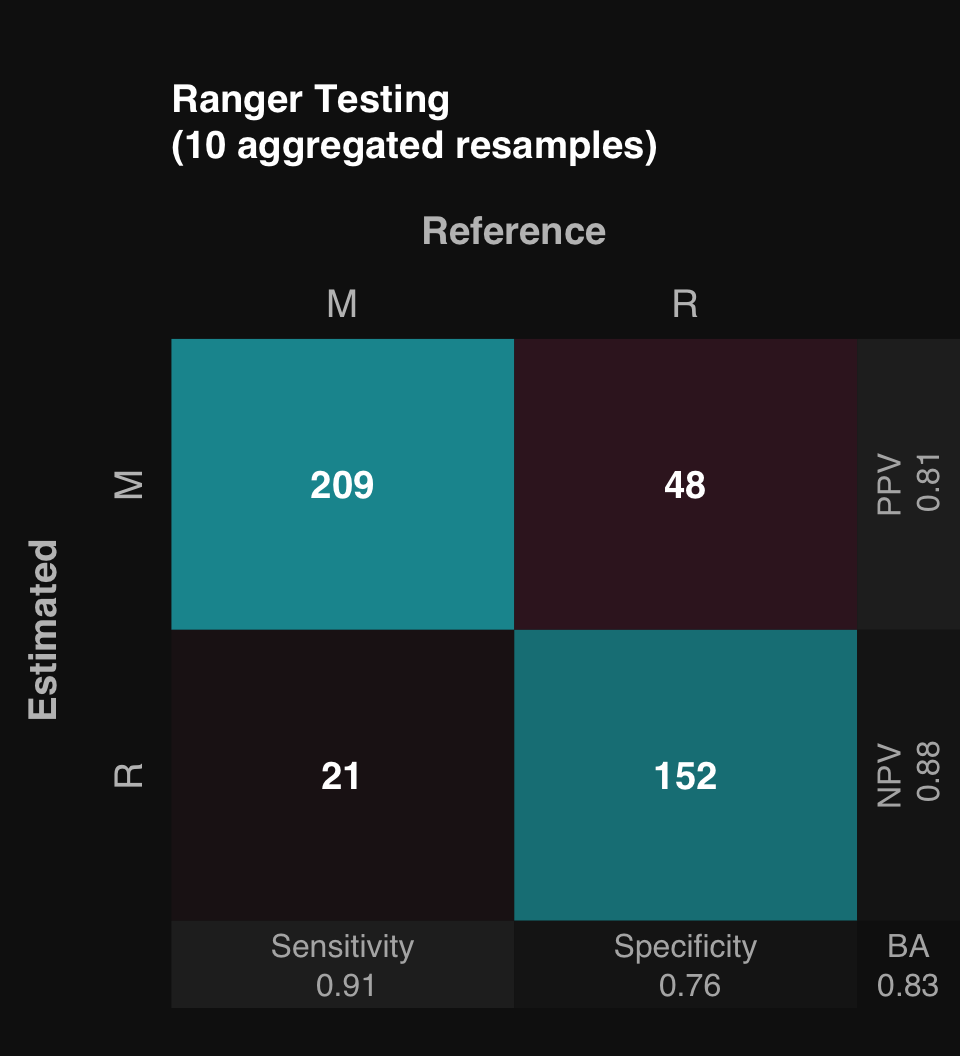
mod$plotROC()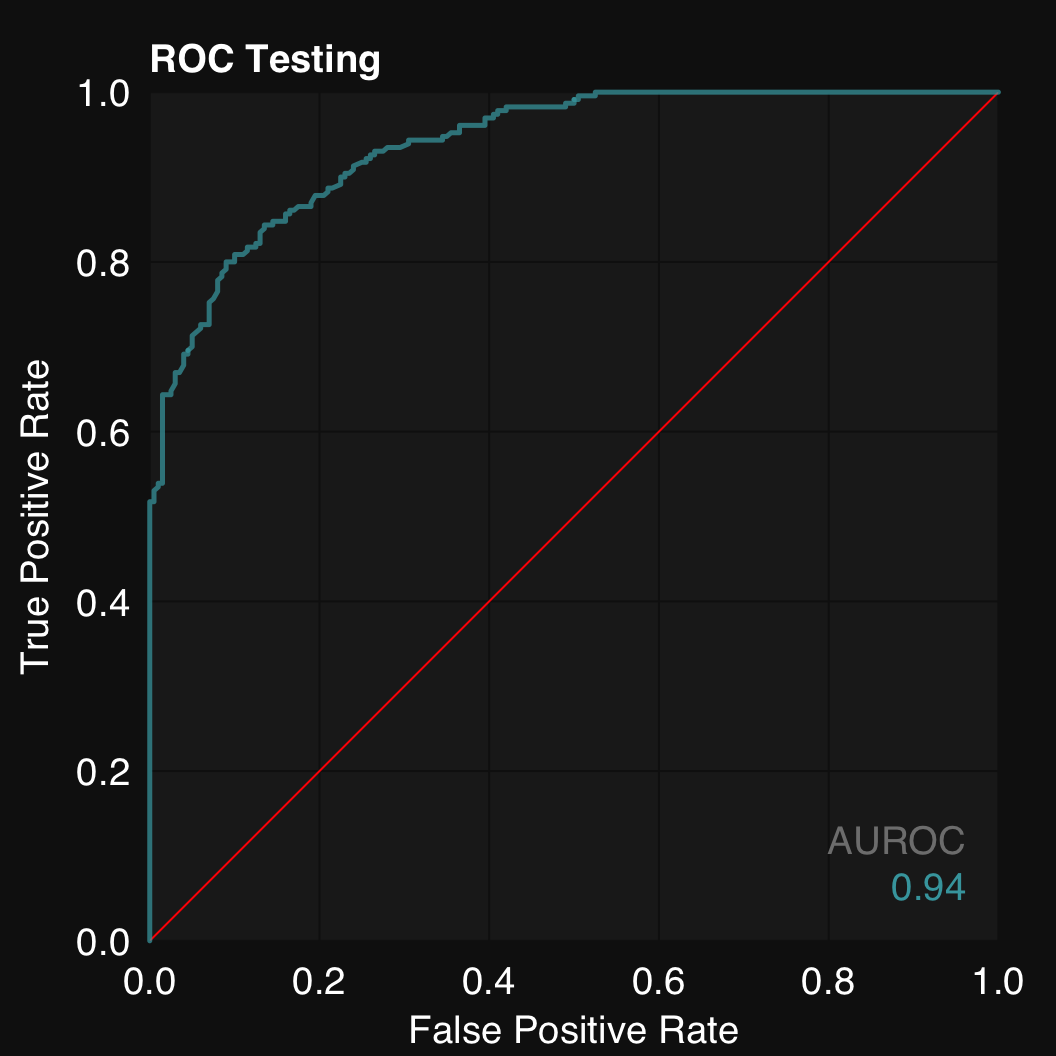
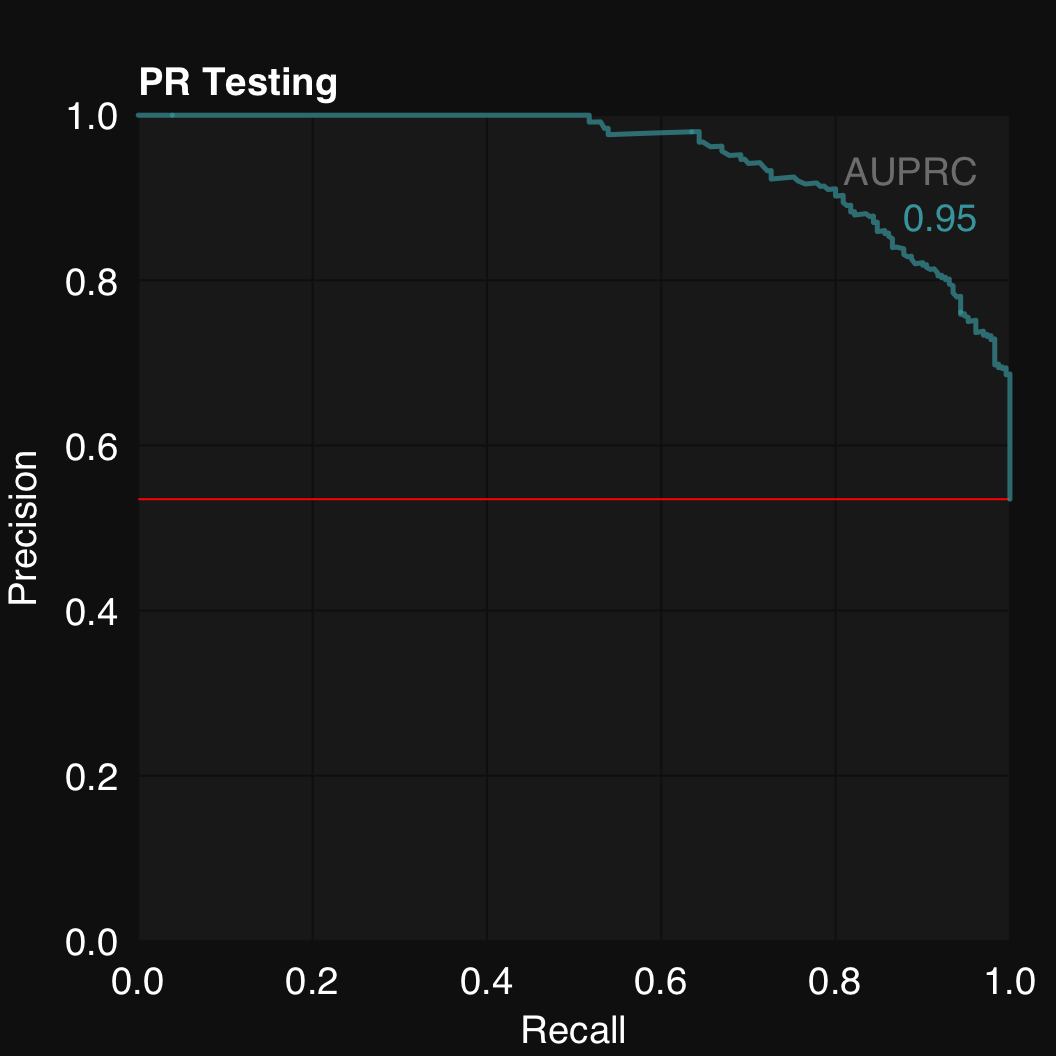
mod$plotPR()